AlphaFold 2, Open Source AI for Protein Structure Prediction
August 2, 2021
On July 15, a team of scientists published a Nature article, titled “Highly accurate protein structure prediction with AlphaFold.”[1] The article describes how the neural network model developed by Google’s DeepMind can predict protein structures “with atomic accuracy even where no similar structure is known.”[2] In addition, DeepMind has now open-sourced the code for AlphaFold 2, allowing further collaborations for even more accurate protein structure prediction.
A protein can have a highly complex 3D structure through a process called protein folding, and the task of predicting the structure has been “an important open research problem for more than 50 years.”[3] Last year DeepMind entered the research competition CASP14 (14th Critical Assessment of protein Structure Prediction), won the competition, and redesigned AlphaFold to create AlphaFold 2 in December 2020. The CASP competitions, considered as “the Olympics of protein folding,”[4] have been held biennially since 1994, and after the development of AlphaFold 2, some view that the protein folding problem has been essentially solved. DeepMind has successfully improved the prediction accuracy “by incorporating novel neural network architectures and training procedures based on the evolutionary, physical, and geometric constraints of protein structure.”[5]
AlphaFold inspired other research efforts, which led to the publication of another article on July 15, “Accurate prediction of protein structures and interactions using a three-track neural network.”[6] The article by academic researchers describes how their RoseTTAFold model predicted protein structures at the accuracy level close to that of AlphaFold. The model features a three-track network where “information at the 1D sequence level, the 2D distance map level, and the 3D coordinate level is successively transformed and integrated.” With such technology “RoseTTAFold enables solutions of challenging X-ray crystallography and cryo-EM modeling problems, provides insight into protein function in the absence of experimentally determined structures, and rapidly generates accurate models of protein-protein complexes.”
Protein misfolding could lead to various diseases and disorders, and thus the availability of computational tools that provide insight into protein folding is significant to drug discovery and development. The prediction models, together with experimental techniques, are expected to help better understand the causes of diseases and design compounds that could effectively treat the diseases.
In terms of patent protection, London-based DeepMind filed three PCT International Applications with the same title “Machine Learning for Determining Protein Structures” on September 16, 2019, claiming priorities to the same three U.S. provisional applications filed in September and November 2018.
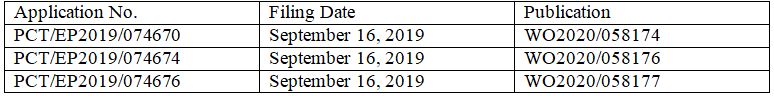
U.S. Provisional Applications:
No. 62/734,757 filed September 21, 2018
No. 62/734,773 filed September 21, 2018
No. 62/770,490 filed November 21, 2018
WO2020/058174 includes claims to a prediction method, a system, and computer storage media. Claim 1 is as follows.
1. A method performed by one or more data processing apparatus for determining a final predicted structure of a given protein, wherein the given protein includes a sequence of amino acids, wherein a predicted structure of the given protein is defined by values of a plurality of structure parameters, the method comprising:
generating a plurality of predicted structures of the given protein, wherein generating a predicted structure of the given protein comprises:
obtaining initial values of the plurality of structure parameters defining the predicted structure;
updating the initial values of the plurality of structure parameters, comprising, at each of a plurality of update iterations:
determining a quality score characterizing a quality of the predicted structure defined by current values of the structure parameters, wherein the quality score is based on respective outputs of one or more scoring
neural networks which are each configured to process: (i) the current values of the structure parameters, (ii) a representation of the sequence of amino acids of the given protein, or (iii) both; and
for one or more of the plurality of structure parameters:
determining a gradient of the quality score with respect to the current value of the structure parameter; and
updating the current value of the structure parameter using the gradient of the quality score with respect to the current value of the structure parameter; and determining the predicted structure of the given protein to be defined by the current values of the plurality of structure parameters after a final update iteration of the plurality of update iterations; and
selecting a particular predicted structure of the given protein as the final predicted structure of the given protein.
The prediction method of Claim 1 generates multiple predicted structures of a given protein, conducts certain calculations, and at the end selects a particular predicted structure of the given protein as the final predicted structure. The calculations involve obtaining initial values of structural parameters defining the predicted structure and updating the values. The updating process includes the following determining process using neural networks (emphasis added):
“determining a quality score characterizing a quality of the predicted structure defined by current values of the structure parameters, wherein the quality score is based on respective outputs of one or more scoring neural networks which are each configured to process: (i) the current values of the structure parameters, (ii) a representation of the sequence of amino acids of the given protein, or (iii) both”
Claim 1 therefore recites the general functions of the neural networks, but does not recite any specific architectures of neural networks. Thus, similar to Ed Garlepp’s discussion on unique disclosure issues with AI, the neural network is treated more like a “black box” in the claim, although DeepMind was presumably working to develop novel network architectures. This claim is a good example of the balance needed by patent practitioners when drafting claims that involve a neural network.
We note the PCT application was filed well before DeepMind conducted more extensive studies in CASP14, facing the challenge to model various unknown protein structures provided in May-August 2020. During the pandemic the team worked on predicting the structure of SARS-CoV-2 Orf8, one of the coronavirus proteins. In view of the serious circumstances DeepMind was sharing the findings and publishing the results as they were obtained. The patent strategy at DeepMind might have shifted toward an open strategy through such work, which resulted in the recent publication of the details of their technology with the source code being made available under an open-source license.
We look forward to tracking the prosecution of this patent as well as the general evolution of this technology.
[1] Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature https://doi.org/10.1038/s41586-021-03819-2 (2021).
[2] Id., Abstract.
[3] Id.
[4] DeepMind (2020). AlphaFold: The making of a scientific breakthrough [Video]. YouTube. https://www.youtube.com/watch?v=gg7WjuFs8F4
[5] Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature https://doi.org/10.1038/s41586-021-03819-2 (2021).
[6] M. Baek et al., Science 10.1126/science.abj8754 (2021).